Classification¶
A classification example explained by the usage of SALSA package on the Ripley data set. The SALSA package provides many different options for stochastically learning a classification model.
This package provides a function salsa and explanation on SALSAModel which accompanies and complements it. The package provides full-stack functionality including cross-validation of all model- and algorithm-related hyperparameters.
Knowledge agnostic usage¶
-
salsa(X, Y[, Xtest])¶ Create a linear classification model with the predicted output
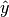 :
: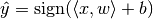
based on data given in
Xand labeling specified inY. Optionally evaluate it onXtest. Data should be given in row-wise format (one sample per row). The classification model is embedded into the returnedmodelasmodel.output. The choice of different algorithms, loss functions and modes will be explained further on this page.using SALSA, MAT, Base.Test srand(1234) ripley = matread(joinpath(Pkg.dir("SALSA"), "data", "ripley.mat")) model = salsa(ripley["X"], ripley["Y"], ripley["Xt"]) # --> SALSAModel(...) @test_approx_eq_eps mean(ripley["Yt"] .== model.output.Ytest) 0.89 0.01
-
salsa(mode, algorithm, loss, X, Y, Xtest) Create a classification model with the specified choice of algorithm, mode and loss function.
Parameters: - mode –
LINEARvs.NONLINEARmode specifies whether to use a simple linear classification model or to apply the Nyström method for approximating the feature map before proceeding with the learning scheme - algorithm – stochastic algorithm to learn a classification model, e.g.
PEGASOS,L1RDAetc. - loss – loss function to use when learning a classification model, e.g.
HINGE,LOGISTICetc. - X – training data (samples) represented by
MatrixorSparseMatrixCSC - Y – training labels
- Xtest – test data for out-of-sample evaluation
Returns: SALSAModelobject.using SALSA, MAT, Base.Test srand(1234) ripley = matread(joinpath(Pkg.dir("SALSA"), "data", "ripley.mat")) model = salsa(LINEAR, PEGASOS, HINGE, ripley["X"], ripley["Y"], ripley["Xt"]) @test_approx_eq_eps mean(ripley["Yt"] .== model.output.Ytest) 0.89 0.01
- mode –
Model-based usage¶
-
salsa(X, Y, model, Xtest) Create a classification model based on the provided model and input data
Parameters: - X – training data (samples) represented by
MatrixorSparseMatrixCSC - Y – training labels
- Xtest – test data for out-of-sample evaluation
- model – model is of type
SALSAModel{L <: Loss, A <: Algorithm, M <: Mode, K <: Kernel}and can be summarized as follows (with default values for named parameters):
mode::Type{M}: mode used to learn the model: LINEAR vs. NONLINEAR (mandatory parameter)algorithm::A: algorithm used to learn the model, e.g. PEGASOS (mandatory parameter)loss_function::Type{L}: type of a loss function used to learn the model, e.g. HINGE (mandatory parameter)kernel::Type{K} = RBFKernel: kernel used in NONLINEAR mode to compute Nyström approximationglobal_opt::GlobalOpt = CSA(): global optimization techniques for tuning hyperparameterssubset_size::Float64 = 5e-1: subset size used in NONLINEAR mode to compute Nyström approximationmax_cv_iter::Int = 1000: maximal number of iterations (budget) for any algorithm in training CVmax_iter::Int = 1000: maximal number of iterations (budget) for any algorithm for final trainingmax_cv_k::Int = 1: maximal number of data points used to compute loss derivative in training CVmax_k::Int = 1: maximal number of data points used to compute loss derivative for final trainingonline_pass::Int = 0: if > 0 we are in the online learning setting going through the entire datasetonline_passtimesnormalized::Bool = true: normalize data (extracting mean and std) before passing it to CV and final learningprocess_labels::Bool = true: process labels to comply with binary (-1 vs. 1) or multi-class classification encodingtolerance::Float64 = 1e-5: the criterion is evaluated for early stopping (online_pass==0)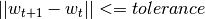
sparsity_cv::Float64 = 2e-1: sparsity weight in the combined cross-validation/sparsity criterion used for the RDA type of algorithmsvalidation_criterion = MISCLASS(): validation criterion used to verify the generalization capabilities of the model in cross-validation
Returns: SALSAModelobject withmodel.outputof typeOutputModelstructured as follows:dfunc::Function: loss function derived from the type specified inloss_function::Type{L}(above)alg_params::Vector: vector of model- and algorithm-specific hyperparameters obtained via cross-validationX_mean::Matrix: row (vector) of extracted column-wise means of inputXifnormalized::Bool = trueX_std::Matrix: row (vector) of extracted column-wise standard deviations of inputXifnormalized::Bool = truemode::M: mode used to learn the model: LINEAR vs. NONLINEARw: found solution vector (matrix)b: found solution offset (bias)
- X – training data (samples) represented by
using SALSA, MAT, Base.Test
srand(1234)
ripley = matread(joinpath(Pkg.dir("SALSA"), "data", "ripley.mat"))
model = SALSAModel(NONLINEAR, R_L1RDA(), HINGE, global_opt=CSA())
model = salsa(ripley["X"], ripley["Y"], model, ripley["Xt"])
@test_approx_eq_eps mean(ripley["Yt"] .== model.output.Ytest) 0.895 0.01